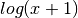skg.cluster1d¶
A one-dimensional clustering algorithm that is useful for many applications. It can be used to decode UART packets and seed iterative minimizers for sinusoidal data.
The algorithm is very simple, relying on one sort, two diffs, and either a max or partition of the data. The sort makes the asymptotic complexity increase as O(n log(n)), but with a remarkably small load factor.
This module is not exactly a fitting rougine, although it is used by
sin_fit, so it is not exported through the main
skg namespace.
Functions
cluster1d(x[, absolute, dscale, sensitivity]) |
Perform a 1-D clustering on the data. |
dscale_log(x) |
Perform an in-place log-scaling of the input, adjusted so that zeros are handled correctly. |
-
skg.cluster1d.cluster1d(x, absolute=True, dscale=None, sensitivity=0)[source]¶ Perform a 1-D clustering on the data.
Data is assumed to be sorted, either increasing or decreasing monotonically. All data will be treated as a raveled 1-D array.
Parameters: - x (array-like) – The data points. Treated as a one-dimensional raveled array. Must be sorted.
- absolute (bool, optional) – Whether or not to introduce an absolute scaling term to the edge length computation. The term will determine whether cluster boundaries are subject to minute fluctuations in the data. It is generally a good idea to have this flag turned on (it is by default).
- dscale (callable, optional) – A function that accepts an array as input, to be applied in-place to the edges lengths before partitioning. This callable will adjust the location of the partition, and therefore what is considered to be a cluster boundary. Useful choices are None (no-op), dscale_log.
- sensitivity (int, optional) – The number non-maximal second-order edges to consider. With
sensitivity=0orsensitivity=1, only the largest gaps are considered. Forsensitivity=2, the second largest category of gaps will be included.
Returns: q – An array of indices of the start of each cluster. The first element is always zero. The indices are always monotonically increasing.
Return type: Notes
q is intended to be used as the index argument to
numpy.ufunc.reduceat. It can also be used as the input tonumpy.splitby stripping off the leading element:q[1:]. Use the functions inskg.utilto slice and dice the indices further.Although funcionally identical, it is likely that
sensitivity=0is slightly more efficient thansensitivity=1.References
- Currently none. This function, is entirely the work of the author. A peer reviewed paper is currently in the works.